
Ivo Hofacker
About
The Hofacker group develops computational methods for the prediction and analysis of RNA structure and makes these available in the ViennaRNA package. The group has contributed novel algorithms for diverse problems in RNA secondary prediction, including RNA-RNA interaction prediction, simulation of RNA folding kinetics, genome-wide identification of functional RNA structures, and design of functional RNA structures. Tertiary structure prediction is being addressed in a coarse-grained, helix-centered, model that can incorporate experimental data e.g. from SAXS experiments.
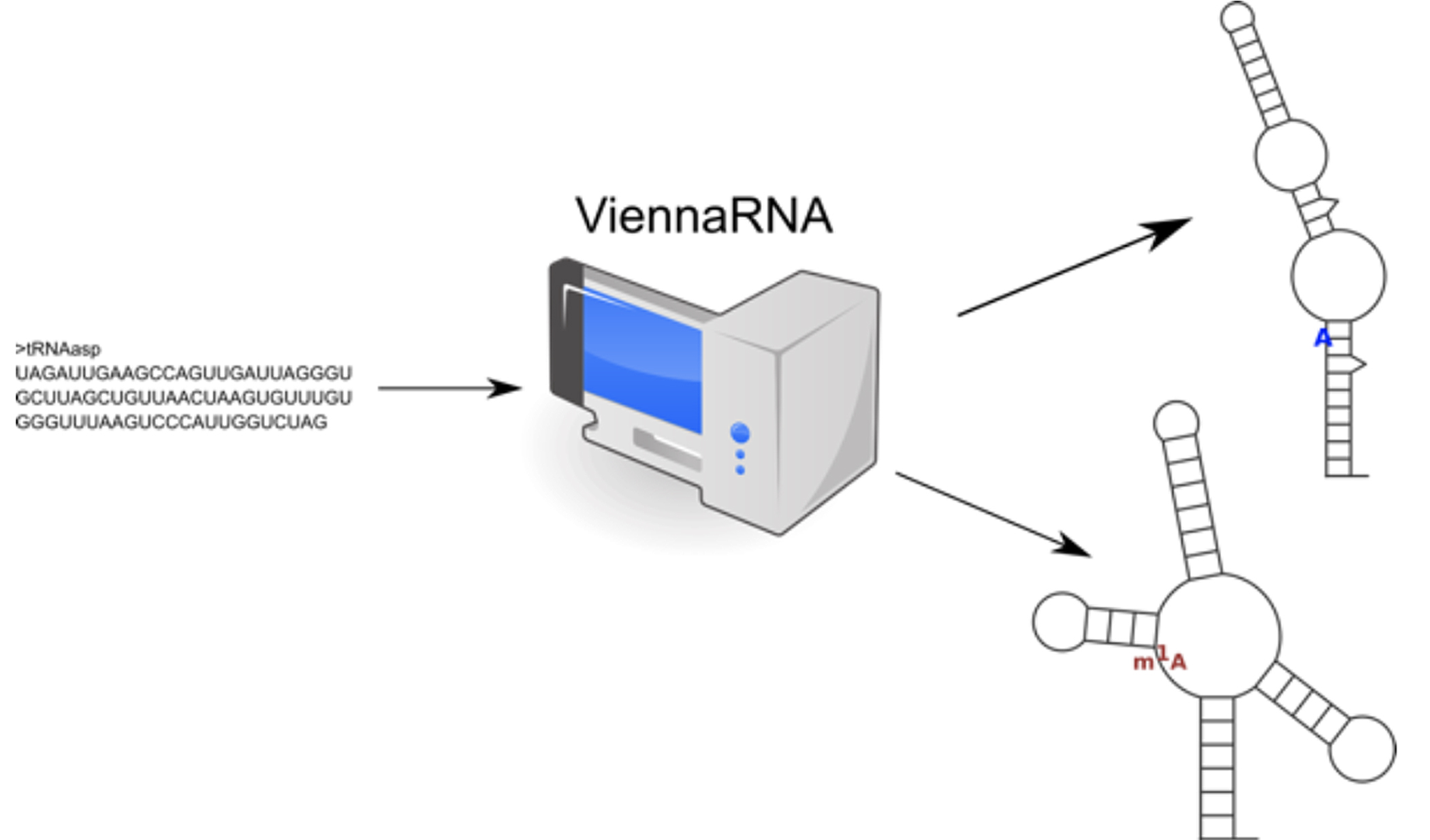
Within this SFB the Hofacker group will study how modifications impact RNA structure, folding dynamics, and interactions. In order to be able to predict these effects, RNA folding algorithms will be adapted to support modified nucleotides, including the estimation of nearest neighbor energy parameters for these modified nucleotides. The resulting modification-aware folding algorithms can be will be applied to systems studied in partner groups, such as tRNAs and their fragments (Schaefer, Villardo, Rossmanith) or Inosine containing mRNAs (Jantsch, Bernecky).
Phone
+43 1 4277 52738
E-mail
ivo@tbi.univie.ac.at
University of Vienna
Institute for Theoretical Chemistry
Währinger Straße 17, 3rd floor, room 312
1090 Wien. Austria
